

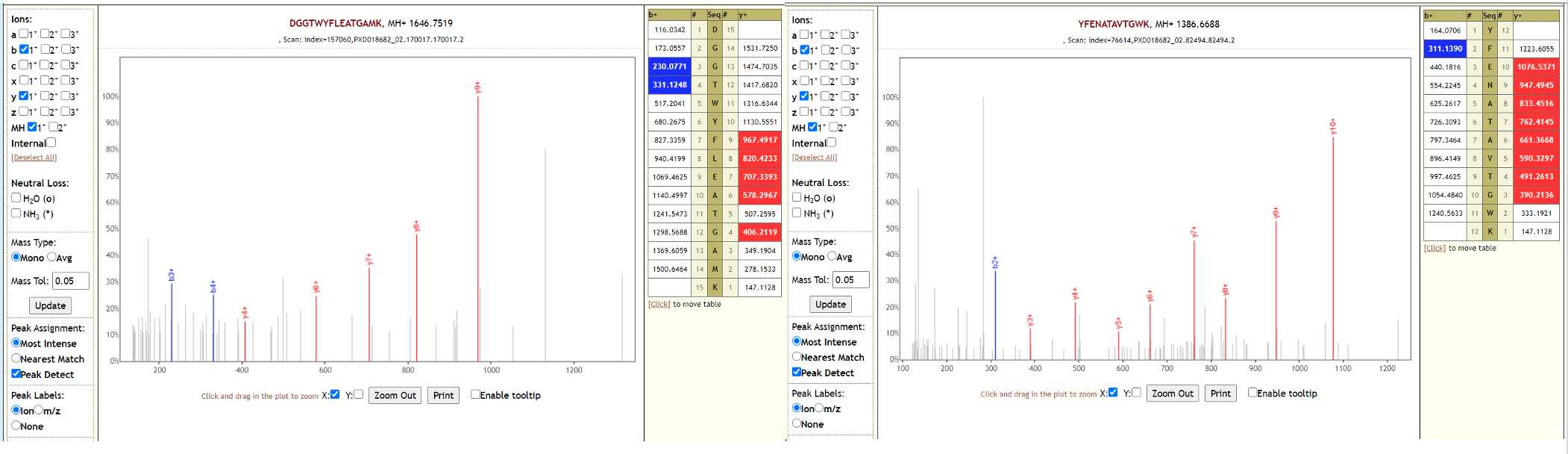
The reliability of all statistical metrics has been validated in detail using the complex Pyrococcus furiosus standard with. Here we present PeptideShaker Online, a user-friendly web-based framework for the identification of mass spectrometry-based proteomics data, from raw file conversion to interactive visualization of the resulting data. In addition to these identification reliability measures, PeptideShaker also provides confident modification site inference using the latest localization methods (Supplementary Note 1). This filter for specificity and sensitivity includes interactive graphs providing immediate feedback on the values of FDR and FNR for PSMs, peptides and proteins at any chosen threshold. Furthermore, as well as providing false discovery rates (FDRs) at the PSM, peptide and protein levels, PeptideShaker calculates reliable false negative rates (FNRs), providing the user with a novel and highly useful interface to filter results according to an FDR-versus-FNR cost-benefit rationale that has so far been absent from proteomics. PeptideShaker provides statistical confidence estimates for each peptide and protein, taking into account protein inference issues (13). To identify peptides and proteins PeptideShaker uses the target-decoy search strategy (10) to estimate posterior error probabilities and uses these to unify the peptide-to-spectrum match (PSM) lists of different search engines, thus increasing the confidence and sensitivity of hits compared with single-search-engine processing (11,12). Importantly, PeptideShaker can work with the combined output of multiple identification algorithms (Fig.
#Peptideshaker software#
Here we describe PeptideShaker (), a proteomics informatics software that can be used at any stage in the proteomics data cycle for the analysis and interpretation of primary data, enabling data sharing and dissemination and re-analysis of publicly available proteomics data.
To maximize the value of public proteomics data, reuse and repurposing must become straightforward, allowing the completion of the proteomics data cycle. There is a pressing need for a user-friendly, open source tool that empowers users to carry out state-of-the-art proteomics data analysis at any stage in the data life cycle (8,9). However, proteomics data processing and (re-)analysis currently remain far from routine practice. Importantly, repository data sharing has enabled the first high-profile studies in which the repurposing of publicly available proteomics data has revealed new biological insights (6,7). Widespread data sharing via publicly accessible repositories, such as PRIDE (1), has now become standard practice, aided by robust user-oriented tools for data submission (2,3) and inspection (4) and bolstered by the advent of the ProteomeXchange initiative (5). Due to resource limitations, the maximum number of concurrent users is set to five and uploading and deleting of files is not supported.Mass spectrometry (MS)-based proteomics is commonly used to identify and quantify the hundreds to thousands of proteins that are present in complex biological samples. DemoĪ fully functional demo is available to test PeptideShaker Online. Start using PeptideShaker Onlineīefore using PeptideShaker Online, please read Start using PeptideShaker Online.
#Peptideshaker how to#
Step-by-step instructions on how to deploy PeptideShaker Online on your own web server are available here: PeptideShaker Online Setup. Note that using a remote Galaxy server includes sending data over the internet which may not be recommended if the data is senstive or requires extra protection. We recommend that both the Galaxy server and the Tomcat server are located on the same machine or in the same network for both performace and security reasons. Galaxy server (local (recommended) or external).The setup of PeptideShaker Online requires access to the following: If you use PeptideShaker Online as part of a publication please include this reference.
#Peptideshaker full size#
(Click on figure to see the full size version) PeptideShaker Online consists of two main components: Galaxy-based backend and a web application as a frontend. PeptideShaker Online is a user-friendly web-based framework for the identification of mass spectrometry-based proteomics data.


 0 kommentar(er)
0 kommentar(er)
